Futuyma, Kirkpatrick (2023) Evolution.
fifth edition
In the
previous blog
I discussed
Bergstrom and Dugatkin Evolution, third and first edition.
Today I continue my investigation of 'junk DNA' in the Evolution textbooks
with a textbook by Douglas Futuyma and
Mark Kirkpatrick (2023)
Evolution also published this year.
Although "junk DNA" does not occur in the index, on page 86 (chapter
'Mutation and variation'), the authors state: "In humans for example, 98% of
the DNA does not code for any gene product." Note: they do not say
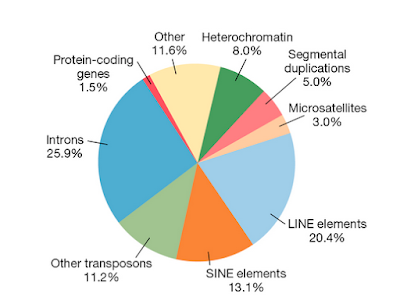
'protein product', but 'gene product'. However, the capture of figure 10.13
states: "Less than 2% is devoted to protein-coding sequences." (p.274).
So, if that is what they mean by 'gene product' the 98% is OK. They explain these matters in an excellent and up-to-date chapter about genes and
genomes (chapter 10). On page 281 the authors ask:
"Does that mean the 98% of our genome that is noncoding is actually
junk? We are still far from having a clear answer to this fundamental
question. (...) some of the resulting "junk" now plays key roles in
regulating gene expression, and the cell's metabolism has coevolved with
the total quantity of DNA in the nucleus. (...) Like an addict and his
drug, eukaryotes may not be able to break their dependence on a bloated
genome. But their is good news in this story. When ancient eukaryotes
acquired large amounts of of noncoding DNA, it opened new options for
the evolution of gene regulation. That, in turn, may have enables the
origin of complex life-forms, including ourselves." (p.281)
In the end-of-chapter section called 'What We Don't Know' (by the
way, a nice feature!):
"Also debated is the fraction of the eukaryotic genome that has a
function. One large-scale study estimated that 80% of the human genome
has a function." [the reference is the
ENCODE publication
in Nature, 2012]. "That estimate, however, has been criticized as
far too high, and many evolutionary biologists would agree that perhaps
only about 10% of our genome has a definite function" (p.281).

|
Figure 10.13. (p.274). Adapted from Gregory (2005)
|
This seems a fair and correct description of the status of the scientific evidence. What I miss in figure 10.13 is the difference between functional
and non-functional DNA (perhaps an unreasonable demand!). The functional sequences outside protein coding
sequences will be hidden in the 98%. 'Functional RNA' is not present in
the book. One can find a comprehensive treatment of functional and
nonfunctional RNA in Moran (2023) (see
my blog
26 June).
"Alternative splicing is a major mechanism used by eukaryotes to
increase organismal complexity " (p.274).
Is it really 'a major mechanism'? This is a controversial statement because there is no quantitative estimate of
its importance. Futuyma and Kirkpatrick do not mention non-coding tRNA (transfer RNA), but ribosomal RNA
(rRNA) is present (p.269). However, both are not introduced as good
examples of non-coding DNA (they do not code for proteins but are
functional).
Note [3] about Futuyma, 1st edition, 1979
Nicholas Barton et al (2007) Evolution
Nicholas Barton et al (2007) Evolution has a very good
discussion of junk DNA, selfish DNA, C-value, non-coding DNA. Fortunately,
they also pay attention to the disadvantages of a big genome (p.597). For
example, in insects metamorphosis requires rapid cell division and is
harder when massive amounts of DNA must be replicated. It shows that
natural selection can downsize large genomes. Which is good to know! It
brings the burden of large genomes back into focus. This is important:
transposons can by accident acquire a new function. They do not
give an estimate how often this happens. "Introns are frequently
considered to be junk DNA. However, comparative sequence analysis has
revealed that the sequence of some introns is highly conserved, suggesting
that functional constraints have played a role in evolution." (p.220).
"Overall, about 18% of nucleotides are conserved in introns and
intergenic regions, compared with 72% within exons" (546). "Such studies
suggest that in that in multicellular eukaryotes, at least as much
non-coding as coding sequence is maintained by selection." (p.547). If all
possible alternative splicing possibilities are taken into account,
species such as humans can make millions of different proteins, even
though they each have only 25,000 protein-coding genes. Alternative
splicing provides a significant source of novelty for diversification"
(p.221). This is regarded as controversial by some. Transposable elements have sometimes been co-opted to aid
their host (p.598). Sometimes pseudogenes acquire new functions.
These views contrast with those of Laurence Moran (2023). Further research is necessary.

|
|
|
|
|
|
|
|
|
Freeman, Herron (2007) Evolutionary Analysis
The most recent edition of Freeman and Herron is the fifth edition (2013). I don't have that edition, I used Evolutionary Analysis 4th edition (2007). There is
no 'junk DNA' and no 'non-coding DNA' in the index. Unexpectedly and
paradoxically, transposons –a prime example of selfish genetic
elements– are discussed in chapter 15 'Phylogenomics and the molecular basis of ADAPTATION'.
But first, read this stunning remark (remember, this book was published before
ENCODE 2012):
"In humans only about 1.2% of the genome codes for proteins."
(p.576)
They do not comment on this remarkable statement. It is an isolated statement from an unknown source. However, they do state
that the "extra" DNA responsible for the C-value paradox consists of
transposable elements: "In the human genome, for example, over 44% of the
DNA present is derived from transposable elements (p.576). What about
the remaining 54%? They do not tell. Unknown?
Fortunately, the authors discuss the burden of these genomic parasites. It costs the cell time, energy and resources to replicate a genome with a lot of transposons (p.577). Funny remark:
transposons are present in the genome in "often appallingly large
numbers"! Such an emotional remark is really funny for a textbook! Good to
know: transposons are not 100% non-coding DNA, because they
encode the enzyme transposase. They have further important
information: defense mechanisms against transposons (!), and:
"work by John Moran (!) (1999) suggested that transposition events in
eukaryotes may occasionally result in mutations that confer a
fitness benefit." (p.581-583). They conclude:
"Even though most transposable elements function as genomic parasites and
most transposition events result in deleterious mutations, it is
increasingly clear that at least some transposition events result in
important new genes or other changes that have a positive impact on the
fitness of organisms." (p.584).
My conclusion: there is
no 'junk DNA' and no 'non-coding DNA' in the index, and more puzzling, there is also no discussion of introns and splicing.
That is a serious omission for an evolution textbook. The origin of introns
is a longstanding evolutionary mystery. However, they have interesting
things to say about transposable elements. According to Laurence Moran 37%
of our genome consists of introns and according to Futuyma, Kirkpatrick
(2023): 26% (see figure above). Obviously, without introns Freeman and
Herron don't have a complete overview of non-coding DNA and can't calculate
the sum total of functionless DNA in our genome. Yet, they know that 1.2% of
the genome codes for proteins! I guess that Freeman and Herron are optimistic about the possibility of finding more
useful elements in the uncharted parts of the human genome and therefore
avoid the concept 'junk DNA'. Reasonable.
Strickberger's Evolution Fourth edition 2008
Strickberger's Evolution is a famous evolution textbook. The first edition was published in 1990. The fifth edition appeared in 2013. The most recent edition
I have is the fourth edition (2008) authored by Brian Hall and Benedikt Hallgrimsson (I don't know whether Strickberger participated in this edition). 'Junk DNA' [1] and 'non-coding DNA' are not in
the index. However, 'junk DNA', 'selfish DNA' and 'C-value paradox' are discussed in
the text.
"According to some molecular biologists, many transposable elements and
other forms of repeated sequences contribute little, if any, function to
their host cells. Because the DNA replication process cannot discriminated
between functional and nonfunctional sequences, it replicates any introduced
sequence. Transposon DNA and repeated sequences may therefore perpetuate
parasitically as either "junk" or "selfish" DNA. (p.221).
Important information is present in Box 12.1 'Quantitative DNA measurements'. In connection with the ENCODE project the following paragraph contains intriguing thoughts which I quote in full:
"According to Bird [1995], eukaryotes were able to circumvent such "noise" by a nuclear membrane that separates transcription from protein translation, allowing only translatable messenger RNA sequences to filter into the cytoplasm, and by tightly folding the DNA of functionally unnecessary genes into nontranscribable confirmations, using nucleosomes and their histones. To these transcription-repressing mechanisms, Bird claims that vertebrates added DNA cytosine methylation, formerly used mostly to suppress genomic parasites such as transposons." (p.260). (my bold)
Especially the concept transcription-repressing is intriguing because according to the ENCODE project and Laurence Moran there is pervasive transcription in the cell and most of it is noise! This would disprove the success of transcription-repressing mechanisms? Apparently, the mechanism fails spectacularly.
My own thoughts are that maybe because transcription is restricted to the nucleus and those RNA transcripts are not exported to the cytoplasm, and consequently are not translated, large-scale transcription can be tolerated by the cell. This assumes that protein synthesis is more costly than transcription. I admit that it is still a burden, but the burden has been halved.
In contrast to Freeman, Herron (2007), in this book 'introns', the "Introns early - Introns late-hypothesis" and alternative splicing are present. Interestingly, they describe introns as mobile DNA sequences that can splice themselves out, acting like transposon-like elements.
Brian Hall and Benedikt Hallgrimsson do not favor the concept 'junk DNA'. It is obvious from this remark: "various biologists have been tempted to consider some or many such sequences as forms of "selfish DNA'." (p.262).
Stephen Stearns, Rolf Hoekstra (2005) Evolution, an introduction, second edition, paperback.
Relevant topics are 'jumping genes', 'transposons', 'introns', 'B-chromosomes'. Not found in other textbooks: B-chromosomes are not transcribed and do not contain information vital to the organism, they are genomic parasites (p.362). This fits the definition of junk DNA, although Stearns and Hoekstra do not use the concept. They make an interesting remark about transposons: several mechanisms have evolved to suppress the deleterious effects of active transposons (p.363). I would like to know more about them! "In humans they [transposons] may account for 45% of the genome". "Transposons illustrate genomic conflict between selection favoring mutants that increase the replication rate of transposons and selection favoring the suppression of transposons through stronger replication control." (p.363). They have a chapter about Genomic Conflict. There is no new edition of this textbook.
Mark Ridley (2004) Evolution, 3rd Edition
Non-coding DNA is listed in the index under 'DNA, non-coding' [2]. One relevant paragraph 2.4: 'Large amounts of non-coding DNA exist in some species'. The human genome contains 5% maybe up to 10% of genes. "The function of non-coding DNA is uncertain. Some biologists argue that it has no function and refer to it as "junk DNA". Others argue that it has structural or regulatory functions." "Most non-coding DNA is repetitive." (p.27). Alternative splicing is mentioned (gene slo), but there is no diagram of exon-intron structure of a gene (!). The existence of genes coding for RNA (rRNA, tRNA) is mentioned in a footnote ("some genes code for RNA" ! p.25). Further information in Chapter 19 'Evolutionary Genomics' is about the evolutionary history of transposable elements ("About 45% of the human genome is derived from transposable elements", p.567). I am a little disappointed, I expected more of Ridley. Please note, that the draft Human Genome sequence was published in 2001. No definitive conclusions possible at that time. There is no new edition.

John Archibald (2018) 'Genomics: A Very Short Introduction', Oxford University Press, 135 pages, has a succinct description of the ENCODE project in the paragraph "Jumping genes and 'junk' DNA" (p.50-53): "The ENCODE project's broadest and most controversial claim is that 80 per cent or more of the human genome has a biochemical function. "
Peter Skelton 'EVOLUTION. A biological and palaeontological approach' (1993, 1994, 1996)
Initially, I ignored this book because I did not expect it would contain junk DNA. Surprise. In chapter 3: Heredity and Variation, the C-value paradox is explained and illustrated with the well-known genome size diagram of various groups of organisms (Fig. 3.5). It immediately stands out that all salamanders and lungfish have bigger genomes than all mammals, birds and reptiles. The C-value paradox is explained by differences in the amount of various repetitive sequences, and polyploidy. Bats and birds have a high metabolic rate and their genome size is lower than other mammals (p.84). lntrons were discovered in 1977. There is a diagram (Fig. 3.9) of gene structure (intron-exon structure). This figure shows introns with smaller sizes than exons. Unfortunately, students get the false impression that these are the right proportions. However, introns are generally larger than exons. Alternative splicing is described (p.90). Transposons are explained (p.92). Conclusion: despite this book appeared before the publication of the human genome in 2001, the ingredients of 'junk DNA' are present. So, it doesn't matter that the word 'junk DNA' isn't used.
Conclusion:
The word 'junk DNA' is absent in the index and in the text of 4 of the 9 textbooks I investigated. However, if 'junk DNA' is not in the index of a textbook, it always pays to search for transposons, introns, jumping genes, selfish DNA, pseudogenes or C-value paradox. This review of evolution textbooks is not exhaustive (I could not check all editions of all textbooks). Those listed here are the most interesting and give sometimes additional useful insights and different points of view. Some are pre-2012, some post-2012, but all except Skelton are post-2001. In general authors know that less than 2% of our DNA codes for proteins, but are not sure about the rest. I agree. Nobody can claim to know exactly how much of our genome is useless junk. Even assuming that 90% of our genome is junk, there is no definitive answer to the question why is there so much junk in our genome, and why it hasn't been eliminated.
Not discussed here is John Parrington (2017) 'The Deeper Genome. Why there is more to the human genome than meets the eye' (OUP paperback). This is a must read. He gives very interesting examples of beneficial non-coding DNA derived from transposons, and has a point of view other than that of Moran (2023). I hope to blog about it in the future.
Thank you for reading!
Notes
- Later I found 'junk DNA' listed under 'Deoxyribonucleic acid' - "junk DNA" in the index! [30 Jul 23]
- Non-coding DNA is listed in the index under 'DNA, non-coding' [30 Jul 23]
- Futuyma (1979) Evolutonary Biology (1st edition). page 439: "Nonetheless, there is an enormous amount of redundancy
in the genome, and its significance is obscure. In may animals as much
as 60% of the genome seems to consists of short (less than 300
nucleotide pairs) repeated sequences, some present in thousands of, even
a milion, copies". That's all. (personal communication Gerdien de Jong) [1 Aug 23]