
|
|
Bergstrom, Dugatkin 2023 |
In a previous post I discussed Larry Moran (2023) 'What's in Your Genome? 90% of Your Genome Is Junk'. He argued that 90% of our genome is junk and has no function until proven otherwise. What about the textbooks? Is 'junk DNA' discussed in the Evolution textbooks? What do they say?
The latest Evolution textbook was published this month: Bergstrom and Dugatkin (2023) Evolution, third edition. There is no 'junk DNA' in the Index, but non-coding DNA is present and 'junk DNA' is mentioned once in chapter 10: Genome Evolution (page 371). There is a lengthy and excellent discussion of the C-value paradox (differences in genome size do not correlate with organism complexity) and the G-value paradox (multicellular eukaryotes tend to have very similar numbers of protein-coding genes despite large differences in organismal complexity). According to the authors the C-value paradox is solved by the observation that most of the genome is non-coding DNA.
"Nonetheless, the additional genetic material may be co-opted in any number of ways, allowing subsequent evolution by natural selection of complex genome organization".
The solution
of the G-value paradox is that regulatory networks matter more than the
total number of genes. Humans have 2000 transcription factors whereas the
nematode has 500. The authors don't speculate about it, but I'd love to know how many transcription factors
chimpanzees have! Then we would know what a difference they make. They could be redundant.
Bergstrom and Dugatkin don't seem to believe in 'junk DNA':
"the noncoding regions of our genome are extensively transcribed and are heavily involved in regulation of gene expression (Mattick et al, 2010)".
According to Moran, John Mattick is one of the ENCODE leaders and one of the most prominent opponents of junk DNA. The authors could not have read Moran, but they could have encountered strong criticism of the ENCODE conclusions. In line with their adaptionist views, and contrary to the view that most alternative spliced genes are splicing errors, the authors think alternative splicing is important in producing different protein products from the same gene. So, introns can have advantages. Further, they argue that transposition (transposons) can potentially have advantages as well. Because the authors think much noncoding DNA could be useful, they do not take the burden of junk DNA serious. If it is functional, there is no burden. Yet, they write that introns impose substantial fitness costs. They do not attempt to quantify the costs. As a consequence they don't have an idea of the magnitude of nonfunctional DNA and the magnitude of the evolutionary costs of junk.
|
|
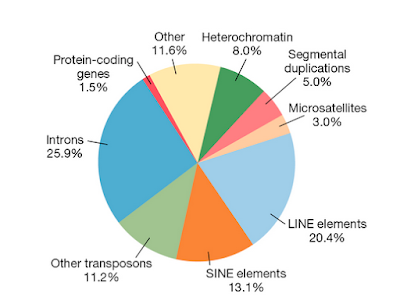
Figure 10.20 Composition of the human genome. Adapted from Gregory (2005) |
In figure 10.20 in chapter 10 protein-coding genes contribute only 1.5% of the total DNA of the human genome. There is a category 'Other' which contributes 11.6% to the total DNA but it is not clear whether it is junk or functional DNA. The authors do not state what the total percentage of junk DNA is. One cannot derive if from this figure because for example the 'Introns' and 'Other transposons' category could contain some useful DNA. My impression is that for the authors junk is an open question. They do not claim that most of our genome is functional.
 |
| Bergstrom, Dugatkin 2012 |
In Bergstrom and Dugatkin (2012) I found interesting remarks about the disadvantages of introns:
"Yet, introns may impose substantial fitness costs as well. First, introns increase the total size of the genome, thereby increasing metabolic costs and decreasing the maximal rate of cell replication. Mutations to the spliceosomal recognition sites can disrupt RNA processing, and thus they can create nonfunctional proteins. Introns also offer refuges for active transposons and other selfish genetic elements that can subsequently cause deleterious mutations." (p.364)
Unfortunately, they do not attempt to calculate the costs. 'junk DNA' is not in the text.
[added 22 Jul 2023]
In a next blog I will discuss other Evolution textbooks.
nice info
ReplyDelete