"Most of the DNA in a genome is junk: The genes exist only as small islands in large oceans of meaningless DNA" (p.544) [1]
 |
| Periannan Senapathy |
These beautiful poetic and prophetic words were written by genome scientist Periannan
Senapathy in 1994 [1]. But he made a more precise claim:
the human genome consists of 90-99.5% junk DNA. This is his most extreme
claim. Elsewhere in his book he writes:
"When we know that greater than 90% of the genome is junk (unused) DNA, and greater than 90% of a gene is introns". (page 146)
He further notes that "Introns are
similar to intergenic junk DNA, except introns occur within individual
genes rather than between genes. The average length of an intron is
approximately ten times longer than the average length of an exon."
Elsewhere he writes: "the proportion of introns in a gene is greater than
90%". He does not refer directly to a specific source for this estimate
[3].
|
|
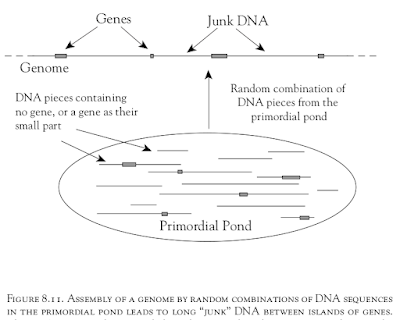
Figure 8.11. Assembly of a genome by random combinations of DNA sequences in the primordial pond leads to long “junk” DNA between islands of genes. |
In 1994 when he published his book no whole genome sequence was
available, let alone a human genome. The sequence of one chromosome of
the eukaryote yeast Saccharomyces cerevisiae
was published in 1994 and two years later the complete genome was
published. Senapathy based his claim on an estimate of evolutionary
biologist and textbook author Douglas Futuyma (1986) that the protein
coding genes of an eukaryote would account for less than 10 percent of the
average genome [2].
Futuyma himself did not use the term 'junk', instead he used 'non-coding
DNA' of unknown function, but Senapathy
interpreted that as junk.
This is very remarkable estimation. This year Laurence Moran wrote a book with the title 'What's in Your Genome? 90% of Your Genome Is Junk' (see my previous blog). So, Senapathy's 90% junk claim predates Moran's claim by almost 30 years. In that sense Moran' was not the first. But, note that Moran's claim is extensively documented, whereas Senapathy's claim is a rough estimate.
Senapathy was aware of the C-value paradox. The C-value is the haploid amount of DNA in a species. The C-value paradox is "the lack of correspondence between C values and the amount of genetic information in the genome." He knew for example that the amount of DNA in each cell of many amphibians and plants is 50 to 100 times larger than that of human beings. He explains the origin of junk DNA and the C-value paradox by random assembly of random DNA in genomes in the 'primordial pond' (Figure 8.11). It follows from his theory that "The genome sizes in various organisms are randomly distributed."
The 90% claim is intimately connected with his theory about the origin of
eukaryotes. I will neither explain nor criticize his theory here because I did
it extensively on my
website. Here I only want to point out that junk DNA and random DNA are the
cornerstone of his theory. He defines junk DNA thus: "junk DNA and introns
do not have any function" and "any change in the junk DNA should, by
nature, be necessarily neutral as far as the function of any gene or the
whole genome is concerned". This is still correct today. He claims on the
basis of statistical analysis of gene sequences in the GenBank database
that "today’s eukaryotic genes are almost random in sequence." (!). It
perfectly fits in his theory and it is consistent with Moran's 90% claim.
Finally, considering his own theory, it is no surprise that Senapathy
fully accepts junk DNA. However, I think it is a biased view. In my next
blog I shall have a look at what the Evolutionary biology
textbooks have to say about 'junk DNA'.
Notes
- Periannan Senapathy (1994) "Independent Birth of Organisms. A New Theory That Distinct Organisms Arose Independently From The Primordial Pond Showing That Evolutionary Theories Are Fundamentally Incorrect". I wrote a lengthy review about the book; I am still working on it.
- His sources are: D. J. Futuyma (1986) Evolutionary Biology, Sinauer Associates, page 48 and database GenBank (Center for Biotechnology Informatics, National Institutes of Health, Bethesda, Maryland).
- In humans, intron lengths contribute 95% of the average gene’s sequence (Venter et al. 2001 'The sequence of the human genome'). That is 7 years after Senapathy's claim. [ 3 Aug 2023 ] According Laurence Moran (2023) 3% of a gene is exon, and 97% is intron. [ 7 Aug 2023 ]
Thanks Bert. High tech biotechnology! I noticed this study demonstrated positive selection and adaptive mutation, adaptation contrary to (random) genetic drift. What I found especially pleasing is the conclusion
ReplyDelete"Instead, natural selection during extended laboratory growth outweighed any deleterious effects of genome disruption and drift..." and:
"rapid adaptation of the minimal cell involved selection on distinct targets"
Hi Bert, sorry for the 'anonymous': blogger makes it very difficult for me to comment with my own name these days.
ReplyDeleteIn 'Evolution of a minimal cell' the authors mean by 'target': the genes that apparently are under the influence of natural selection and change more rapidly and systematically than other genes during the time of the experiment.
What are the key points in your blog post critiquing Periannan Senapathy's 1994 assertions about genetic sequences? Tel U
ReplyDelete