|
|
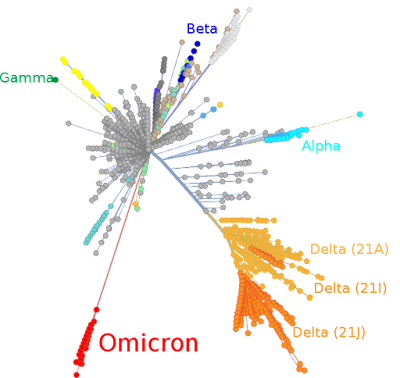
SARS-CoV-2 Omicron variant.
©wikipedia. A picture is worth a thousand words! (click on the image for the fireworks variant!) |
Picture: Omicron and other SARS-CoV-2 variants depicted in a tree scaled radially by genetic distance, derived from Nextstrain on 1 December 2021. The centre of the tree represents the origin of the variants.
How the Omicron Variant Got So Many Scary Mutations So Quickly (Scientific American). Indeed, the Omicron variant has 60 mutations compared to the original Wuhan virus– with 32 in the Spike protein. That's a lot. But what is lacking in the SciAm article is a clear visual display. The image above from wikipedia makes it clear immediately: Omicron is exceptional. Whereas other variants keep accumulating mutations gradually and generate new variants, the Omicron variant seems to have made a giant jump. We don't see intermediates in the evolutionary tree. Omicron is not the result of adding some mutations to -say- the Delta variant. It is not a descendant of Delta. It sets itself apart from all other variants. In the evolutionary tree above it seems the origin of Omicron is situated very close to the first SARS-CoV-2 virus detected in 2019 in Wuhan [1]. It seems to appear out of nowhere, although there is no doubt that it is a SARS-CoV-2 virus [2]. If anything looks like a 'genetically engineered lab escape', it is Omicron, not the original Wuhan virus! (just kidding!) [4]
But how did the Omicron variant got so many mutations so quickly? We don't
know for sure, but the
Scientific American article has a few suggestions. The numerous
changes in the coronavirus's Spike protein could have arisen in an
isolated population or an immunocompromised person. Immunocompromised
patients have severely impaired immunity. A weakened immune system
may result from various causes: due to treatment with drugs that suppress
the immune system, after an organ transplant, due to bone marrow disease,
or during cancer treatment or someone with HIV. In these persons the virus
could stay and replicate for a much longer time than in persons with a
normal immune system. During that time the virus has the opportunity 'to
test' many mutants. A process of mutation and natural selection in the
body of the patient. So, the origin of Omicron could be a single
individual. According to the SciAm article, South African
researchers described an HIV-positive woman who had a SARS-CoV-2 infection
for more than six months! That is long enough to try thousands of mutants.
HIV is the Human Immunodeficiency virus. It attacks the immune
system. The very system that should protect us against viruses!
In
South Africa there are a lot of HIV patients. South Africa is the region
where Omicron was first detected.
This is #37 in the series of Corona updates
Notes
- Omicron ... "descended not from one of the other variants of concern, such as Alpha, Beta or Delta, but from coronavirus that was circulating maybe 18 months ago. ... "The Alpha and Beta variants, first spotted last year in the UK and South Africa respectively, are widely thought to have emerged after long-term infections in patients." from: Why fighting Omicron should include ramping up HIV prevention, The Guardian, 11 Dec 2021
- Omicron is classified as B.1.1.529 (in the Pango classification system [3]), thus has been assigned to the B.1.1 lineage of SARS-CoV-2. See the website cov-lineages.org for a table of all variants. Surprisingly, there are indeed 529 variants in the B.1.1 lineage. Interestingly, the closest variant (with no issue) is: B.1.1.522 (Belgium lineage) from 2020-12-09. That means that B.1.1.529 is separated by 11 months with B.1.1.522. If we include lineages 'with issues' than B.1.1.528 is the closest lineage with the date 2020-12-07, that is also 11 months difference. So, there is a gap of 11 months between B.1.1.529 and its closest relatives. updated 18 Dec 21
- A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology, Nature Microbiology, 15 July 2020 (Proposes the Pango nomenclature system used in cov-lineages.org).
added 18 Dec 21 - “It just came out of nowhere,” says Darren Martin, a computational biologist at the University of Cape Town, South Africa. Nature 28 Jan 22
Sources
- Sarah Wild How the Omicron Variant Got So Many Scary Mutations So Quickly, Scientific American,
-

No comments:
Post a Comment
Comments to posts >30 days old are being moderated.
Safari causes problems, please use Firefox or Chrome for adding comments.