
|
|
corona update #42 |
No evolution without mutation!
Watch SARS-CoV-2's growing number of mutations during the past 2 pandemic years:

|
|
fig. 1. The range of nucleotide mutations of all
SARS-CoV-2 variants
in the past two years (Nextstrain). Captions in the graph are mine. |
Each dot represents one sequenced virus. That means all ≈ 30,000 of the virus' genetic instructions are known. On the vertical axis is the number of mutations. On the horizontal axis is time. Virus variants (clades) have different colours. One thing is clear from this graph: the number of mutations in SARS-CoV-2 has steadily increased from the very first SARS-COV-2 virus in December 2019 until January 2022. There is a linear increase of the number of mutations.
Please note: there are thousands of different
mutations in all SARS-CoV-2 viruses worldwide. But they do not occur in
one individual virus at the same time. The virus has a total of 29,000 –
30,000 nucleotides. Potentially, each nucleotide can be mutated. Or
deleted. So, in theory, up to 30,0000 point mutations are possible. I
don't know how many mutations are possible before the virus breaks down.
All mutations in this graph are relative to the first virus genome from
Wuhan. The number of mutations of the Wuhan sample is set to zero. The
Wuhan sample is at the bottom left corner. When I captured the graph
the highest number was
132 nucleotide mutations.

|
|
fig. 2. A few Omicron samples with the highest number of mutations
(Nextstrain) (numbers in graph are mine) |
In this graph I selected the two Omicron variants 21L, 21K. As an
illustration I added the number of mutations of 4 samples. So far 148
mutations is the maximum. Unfortunately, today I didn't find it anymore in
the Nextstrain database. Today the next highest has 142 mutations (nextstrain). Samples are continually added to Nextstrain and apparently sometimes
deleted.
- Please note there is a bug in the Nextstrain software: in figure 2 the higher number of mutations (132, 148) end up below the lower numbers (122,125).
Not all mutations of a virus are important. So-called 'synonymous mutations' do not change amino acids of the virus protein. But 'non-synonymous mutations' do change amino acids. The non-synonymous mutations could change the function of the protein. And that could change the function of the virus. However, it is no trivial matter to establish what the effect of a specific mutation or combination of mutations (!) is on the behaviour of the virus. The virus could become more infectious, more deadly or produce side-effects in people. For example, the Delta and Omicron variant outcompeted other variants and became dominant. That is difficult to predict, especially with a new virus. It all depends on the interaction of the virus with the human body.
Why such a regular accumulation of mutations?
When I look again at the graphs with the ever increasing mutations, I am still puzzled. Why does it behave in this way? If a virus is successful, and yes this virus is absolutely successful, then why change? If it ain't broke, don't fix it! Or is it successful because it keeps mutating? I think a virus that stops mutating -if it could!- isn't able to keep up with countermeasures of its new host: increasing immunity [1], vaccines, lockdowns, etc.
"Now, here, you see, it takes all the running you can do, to keep in the same place. If you want to get somewhere else, you must run at least twice as fast as that!" (Red Queen's race)
DIY: Do it yourself
Don't believe me, do it yourself! It's fun and educational to experiment with the Nextstrain tool:- Go to https://nextstrain.org/
- choose: SARS-CoV-2 (COVID-19) / Latest global analysis-GISAID data
- default dataset (left panel)= ncov (novel corona virus 2019)
- default Date range = 2019-12-16 tot 2022-01-27
-
Color by: Clade (there are now 23 clades)
-
Tree options: default= 'RECTANGULAR'. I used 'CLOCK' option in
the graphs above. Please note that when the CLOCK layout is chosen, the
number of substitutions per year is displayed: "rate estimate: 23,709
substitutions per year" according to Nextstrain. So, that is 47
mutations in two pandemic years. However, note that individual viruses
can have as many as 3 times more mutations! (142).
- Filter Data: default is nothing. Result: "Showing 3223 of 3223 genomes sampled between Dec 2019 and Jan 2022" (assessed 31 Jan 22).
-
Filter for example by entering 'omicron', then choose 21L
and/or 21K from the drop-down list. Result: "Showing 465 of 3223 genomes sampled between Nov 2021 and Jan 2022." (assessed 31 Jan 22). Active filters are indicated in the
graph.
-
Show branches: toggle ON/OFF for a clearer overview.
- 'zoom to selected' button in the top of the graph panel. (optional)
- Point your mouse to a dot in the graph to get information or click on the dot the get more information about the selected virus sequence.
-
Date range: 'Play' watch the variants coming in to existence and
producing branches.
There is much more to explore! There are many unanswered questions. For example: how many nucleotide mutations of the total number of mutations (%) are Amino Acid mutations? Or explore for example which genes in the virus were mutated early on in the pandemic? And of course: will the maximum number of mutations still increase this year?
Notes
- “The virus has to change to stick around,” says Ben Murrell, an
interdisciplinary virologist at the Karolinska Institute in Stockholm.
The receptor-binding domain, where many of Omicron’s mutations are
concentrated, is an easy target for antibodies, and probably comes under
pressure to change in a long-term infection." Nature 28 Jan 22
Postscript
3 Feb 22
The above graphs suggest a gradual evolution. But Omicron is certainly an exception. At least its origin. “It just came out of nowhere,” says Darren Martin. Intermediate steps are not known. Despite its unknown origin it fits the curve of a linear increase of mutations.
Postscript
4 Feb 22
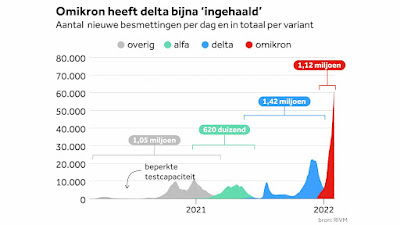 |
| Explosive growth of Omicron variant in the Netherlands (NOS/rivm) |
Previous blogs
-
The exceptional nature of the Omicron variant: a picture is worth a
thousand words!
blog 14 December 2021

No comments:
Post a Comment
Comments to posts >30 days old are being moderated.
Safari causes problems, please use Firefox or Chrome for adding comments.